Example 2: Simple 2D cell signaling model#
We model a reaction between the cell interior and cell membrane in a 2D geometry:
Cyto - 2D cell “volume”
PM - 1D cell boundary (represents plasma membrane)
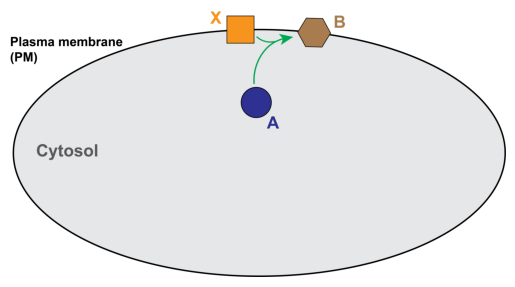
Model from Rangamani et al, 2013, Cell. A cytosolic species, “A”, reacts with a species on the PM, “B”, to form a new species on the PM, “X”. The resulting PDE and boundary condition for species A are as follows:
Similarly, the PDEs for X and B are given by: $\( \frac{\partial{N_X}}{\partial{t}} = D_X \nabla ^2 N_X - k_{on} C_A N_X + k_{off} N_B \quad \text{on} \; \Gamma_{PM}\\ \frac{\partial{N_B}}{\partial{t}} = D_B \nabla ^2 N_B + k_{on} C_A N_X - k_{off} N_B \quad \text{on} \; \Gamma_{PM} \)$
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
img_A = mpimg.imread('axb-diagram.png')
plt.imshow(img_A)
plt.axis('off')
(-0.5, 5830.5, 3140.5, -0.5)
Imports and logger initialization:
import dolfin as d
import sympy as sym
import numpy as np
import pathlib
import logging
import gmsh # must be imported before pyvista if dolfin is imported first
from smart import config, common, mesh, model, mesh_tools, visualization
from smart.units import unit
from smart.model_assembly import (
Compartment,
Parameter,
Reaction,
Species,
SpeciesContainer,
ParameterContainer,
CompartmentContainer,
ReactionContainer,
)
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
logger = logging.getLogger("smart")
logger.setLevel(logging.INFO)
/usr/lib/python3/dist-packages/scipy/__init__.py:146: UserWarning: A NumPy version >=1.17.3 and <1.25.0 is required for this version of SciPy (detected version 1.26.4
warnings.warn(f"A NumPy version >={np_minversion} and <{np_maxversion}"
First, we define the various units for use in the model.
um = unit.um
molecule = unit.molecule
sec = unit.sec
dimensionless = unit.dimensionless
D_unit = um**2 / sec
surf_unit = molecule / um**2
flux_unit = molecule / (um * sec)
edge_unit = molecule / um
Next we generate the model by assembling the compartment, species, parameter, and reaction containers (see Example 1 or API documentation for more details).
# =============================================================================================
# Compartments
# =============================================================================================
# name, topological dimensionality, length scale units, marker value
Cyto = Compartment("Cyto", 2, um, 1)
PM = Compartment("PM", 1, um, 3)
cc = CompartmentContainer()
cc.add([Cyto, PM])
# =============================================================================================
# Species
# =============================================================================================
# name, initial concentration, concentration units, diffusion, diffusion units, compartment
A = Species("A", 1.0, surf_unit, 100.0, D_unit, "Cyto")
X = Species("X", 1.0, edge_unit, 1.0, D_unit, "PM")
B = Species("B", 0.0, edge_unit, 1.0, D_unit, "PM")
sc = SpeciesContainer()
sc.add([A, X, B])
# =============================================================================================
# Parameters and Reactions
# =============================================================================================
# Reaction of A and X to make B (Cyto-PM reaction)
kon = Parameter("kon", 1.0, 1/(surf_unit*sec))
koff = Parameter("koff", 1.0, 1/sec)
r1 = Reaction("r1", ["A", "X"], ["B"],
param_map={"on": "kon", "off": "koff"},
species_map={"A": "A", "X": "X", "B": "B"})
pc = ParameterContainer()
pc.add([kon, koff])
rc = ReactionContainer()
rc.add([r1])
Now we create a circular mesh (mesh built using gmsh in smart.mesh_tools), along with marker functions mf2 and mf1.
# Create mesh
h_ellipse = 0.1
xrad = 1.0
yrad = 1.0
surf_tag = 1
edge_tag = 3
ellipse_mesh, mf1, mf2 = mesh_tools.create_ellipses(xrad, yrad, hEdge=h_ellipse,
outer_tag=surf_tag, outer_marker=edge_tag)
visualization.plot_dolfin_mesh(ellipse_mesh, mf2, view_xy=True)
Info : Meshing 1D...
Info : Meshing curve 1 (Ellipse)
Info : Done meshing 1D (Wall 0.00980047s, CPU 0.009238s)
Info : Meshing 2D...
Info : Meshing surface 1 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0366075s, CPU 0.036696s)
Info : 455 nodes 909 elements
Info : Writing 'tmp_ellipse_[1.0, 1.0]_[0.0, 0.0]_0/ellipses.msh'...
Info : Done writing 'tmp_ellipse_[1.0, 1.0]_[0.0, 0.0]_0/ellipses.msh'
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
ROOT -2025-09-03 20:30:09,777 trame_server.utils.namespace - INFO - Translator(prefix=None) (namespace.py:65)
ROOT -2025-09-03 20:30:09,819 wslink.backends.aiohttp - INFO - awaiting runner setup (__init__.py:138)
ROOT -2025-09-03 20:30:09,819 wslink.backends.aiohttp - INFO - awaiting site startup (__init__.py:145)
ROOT -2025-09-03 20:30:09,821 wslink.backends.aiohttp - INFO - Print WSLINK_READY_MSG (__init__.py:151)
ROOT -2025-09-03 20:30:09,821 wslink.backends.aiohttp - INFO - Schedule auto shutdown with timout 0 (__init__.py:159)
ROOT -2025-09-03 20:30:09,822 wslink.backends.aiohttp - INFO - awaiting running future (__init__.py:162)
Write mesh and meshfunctions to file, then create mesh.ParentMesh object.
mesh_folder = pathlib.Path("ellipse_mesh_AR1")
mesh_folder.mkdir(exist_ok=True)
mesh_file = mesh_folder / "ellipse_mesh.h5"
mesh_tools.write_mesh(ellipse_mesh, mf1, mf2, mesh_file)
parent_mesh = mesh.ParentMesh(
mesh_filename=str(mesh_file),
mesh_filetype="hdf5",
name="parent_mesh",
)
2025-09-03 20:30:09,999 smart.mesh - INFO - HDF5 mesh, "parent_mesh", successfully loaded from file: ellipse_mesh_AR1/ellipse_mesh.h5! (mesh.py:245)
2025-09-03 20:30:10,000 smart.mesh - INFO - 0 subdomains successfully loaded from file: ellipse_mesh_AR1/ellipse_mesh.h5! (mesh.py:258)
Initialize model and solvers.
config_cur = config.Config()
config_cur.solver.update(
{
"final_t": 10.0,
"initial_dt": 0.05,
"time_precision": 6,
}
)
model_cur = model.Model(pc, sc, cc, rc, config_cur, parent_mesh)
model_cur.initialize()
2025-09-03 20:30:16,339 smart.model - WARNING - Warning! Initial L2-norm of compartment PM is 1.119037357695821 (possibly too large). (model.py:1223)
2025-09-03 20:30:16,344 smart.solvers - INFO - Jpetsc_nest assembled, size = (581, 581) (solvers.py:199)
2025-09-03 20:30:16,345 smart.solvers - INFO - Initializing block residual vector (solvers.py:207)
2025-09-03 20:30:16,353 smart.model_assembly - INFO -
╒══════╤═════════════════════════╤═══════════════╕
│ │ Value/Equation │ Description │
╞══════╪═════════════════════════╪═══════════════╡
│ kon │ 1.00×10⁰ µm²/molecule/s │ │
├──────┼─────────────────────────┼───────────────┤
│ koff │ 1.00×10⁰ 1/s │ │
╘══════╧═════════════════════════╧═══════════════╛
(model_assembly.py:371)
2025-09-03 20:30:16,358 smart.model_assembly - INFO -
╒════╤═══════════════╤════════════════╤═══════════════════════╕
│ │ Compartment │ D │ Initial condition │
╞════╪═══════════════╪════════════════╪═══════════════════════╡
│ A │ Cyto │ 1.00×10² µm²/s │ 1.00×10⁰ molecule/µm² │
├────┼───────────────┼────────────────┼───────────────────────┤
│ X │ PM │ 1.00×10⁰ µm²/s │ 1.00×10⁰ molecule/µm │
├────┼───────────────┼────────────────┼───────────────────────┤
│ B │ PM │ 1.00×10⁰ µm²/s │ 0.00×10⁰ molecule/µm │
╘════╧═══════════════╧════════════════╧═══════════════════════╛
(model_assembly.py:371)
2025-09-03 20:30:16,706 smart.model_assembly - INFO -
╒══════╤══════════════════╤═══════════╤════════════╤═════════╤════════════════╤══════════════╕
│ │ Dimensionality │ Species │ Vertices │ Cells │ Marker value │ Size │
╞══════╪══════════════════╪═══════════╪════════════╪═════════╪════════════════╪══════════════╡
│ Cyto │ 2 │ 1 │ 455 │ 845 │ 1 │ 3.14×10⁰ µm² │
├──────┼──────────────────┼───────────┼────────────┼─────────┼────────────────┼──────────────┤
│ PM │ 1 │ 2 │ 63 │ 63 │ 3 │ 6.28×10⁰ µm │
╘══════╧══════════════════╧═══════════╧════════════╧═════════╧════════════════╧══════════════╛
(model_assembly.py:371)
2025-09-03 20:30:16,711 smart.model_assembly - INFO -
╒════╤═════════════╤════════════╤════════════════╤════════════════╕
│ │ Reactants │ Products │ Equation │ Type │
╞════╪═════════════╪════════════╪════════════════╪════════════════╡
│ r1 │ ['A', 'X'] │ ['B'] │ kon*A*X-koff*B │ volume_surface │
╘════╧═════════════╧════════════╧════════════════╧════════════════╛
(model_assembly.py:371)
╒════╤═════════════╤══════╤══════════════════╤═════════════╤══════════════╤════════════════╕
│ │ name │ id │ dimensionality │ num_cells │ num_facets │ num_vertices │
╞════╪═════════════╪══════╪══════════════════╪═════════════╪══════════════╪════════════════╡
│ 0 │ parent_mesh │ 18 │ 2 │ 845 │ 1299 │ 455 │
├────┼─────────────┼──────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 1 │ Cyto │ 32 │ 2 │ 845 │ 1299 │ 455 │
├────┼─────────────┼──────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 2 │ PM │ 39 │ 1 │ 63 │ 63 │ 63 │
╘════╧═════════════╧══════╧══════════════════╧═════════════╧══════════════╧════════════════╛
Save model information to .pkl file and write initial conditions to file.
model_cur.to_pickle('model_cur.pkl')
results = dict()
result_folder = pathlib.Path("resultsEllipse_AR1_loaded")
result_folder.mkdir(exist_ok=True)
for species_name, species in model_cur.sc.items:
results[species_name] = d.XDMFFile(
model_cur.mpi_comm_world, str(result_folder / f"{species_name}.xdmf")
)
results[species_name].parameters["flush_output"] = True
results[species_name].write(model_cur.sc[species_name].u["u"], model_cur.t)
Solve the system until model_cur.t > model_cur.final_t.
tvec = [0]
avg_A = [A.initial_condition]
avg_X = [X.initial_condition]
avg_B = [B.initial_condition]
# Set loglevel to warning in order not to pollute notebook output
logger.setLevel(logging.WARNING)
while True:
# Solve the system
model_cur.monolithic_solve()
# Save results for post processing
for species_name, species in model_cur.sc.items:
results[species_name].write(model_cur.sc[species_name].u["u"], model_cur.t)
dx = d.Measure("dx", domain=model_cur.cc['Cyto'].dolfin_mesh)
int_val = d.assemble_mixed(model_cur.sc['A'].u['u']*dx)
volume = d.assemble_mixed(1.0*dx)
avg_A.append(int_val / volume)
avg_X.append(d.assemble_mixed(X.u["u"]*d.Measure("dx",PM.dolfin_mesh))
/ (d.assemble_mixed(1.0*d.Measure("dx",PM.dolfin_mesh))))
avg_B.append(d.assemble_mixed(B.u["u"]*d.Measure("dx",PM.dolfin_mesh))
/ (d.assemble_mixed(1.0*d.Measure("dx",PM.dolfin_mesh))))
tvec.append(model_cur.t)
# End if we've passed the final time
if model_cur.t >= model_cur.final_t:
break
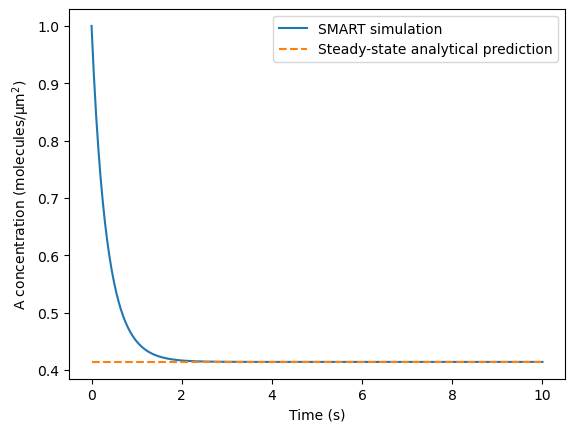
Now we plot the concentration of A in the cell over time and compare this to analytical predictions for a high value of the diffusion coefficient. As \(D_A \rightarrow \infty\), the steady state concentration of A will be given by the positive root of the following polynomial:
Note that in this 2D case, \(SA_{PM}\) is the perimeter of the ellipse and \(vol_{cyto}\) is the area of the ellipse. These can be thought of as a surface area and volume if we extrude the 2D shape by some characteristic thickness.
plt.plot(tvec, avg_A, label='SMART simulation')
plt.xlabel('Time (s)')
plt.ylabel('A concentration $\mathrm{(molecules/μm^2)}$')
SA_vol = 4/(xrad + yrad)
root_vals = np.roots([-kon.value,
-kon.value*X.initial_condition*SA_vol - koff.value + kon.value*A.initial_condition,
koff.value*A.initial_condition])
ss_pred = root_vals[root_vals > 0]
plt.plot(tvec, np.ones(len(avg_A))*ss_pred, '--', label='Steady-state analytical prediction')
plt.legend()
percent_error_analytical = 100*np.abs(ss_pred-avg_A[-1])/ss_pred
assert percent_error_analytical < 0.1,\
f"Failed test: Example 2 results deviate {percent_error_analytical:.3f}% from the analytical prediction"
Plot A concentration in the cell at the final time point.
visualization.plot(model_cur.sc["A"].u["u"], view_xy=True)
# also save to file for comparison visualization in the end
meshimg_folder = pathlib.Path("mesh_images")
meshimg_folder = meshimg_folder.resolve()
meshimg_folder.mkdir(exist_ok=True)
meshimg_file = meshimg_folder / "ellipse_mesh_AR1.png"
# Put all images in a dictionary for easy recovery later
images = {}
images[1] = visualization.plot(model_cur.sc["A"].u["u"], view_xy=True, filename=meshimg_file)
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
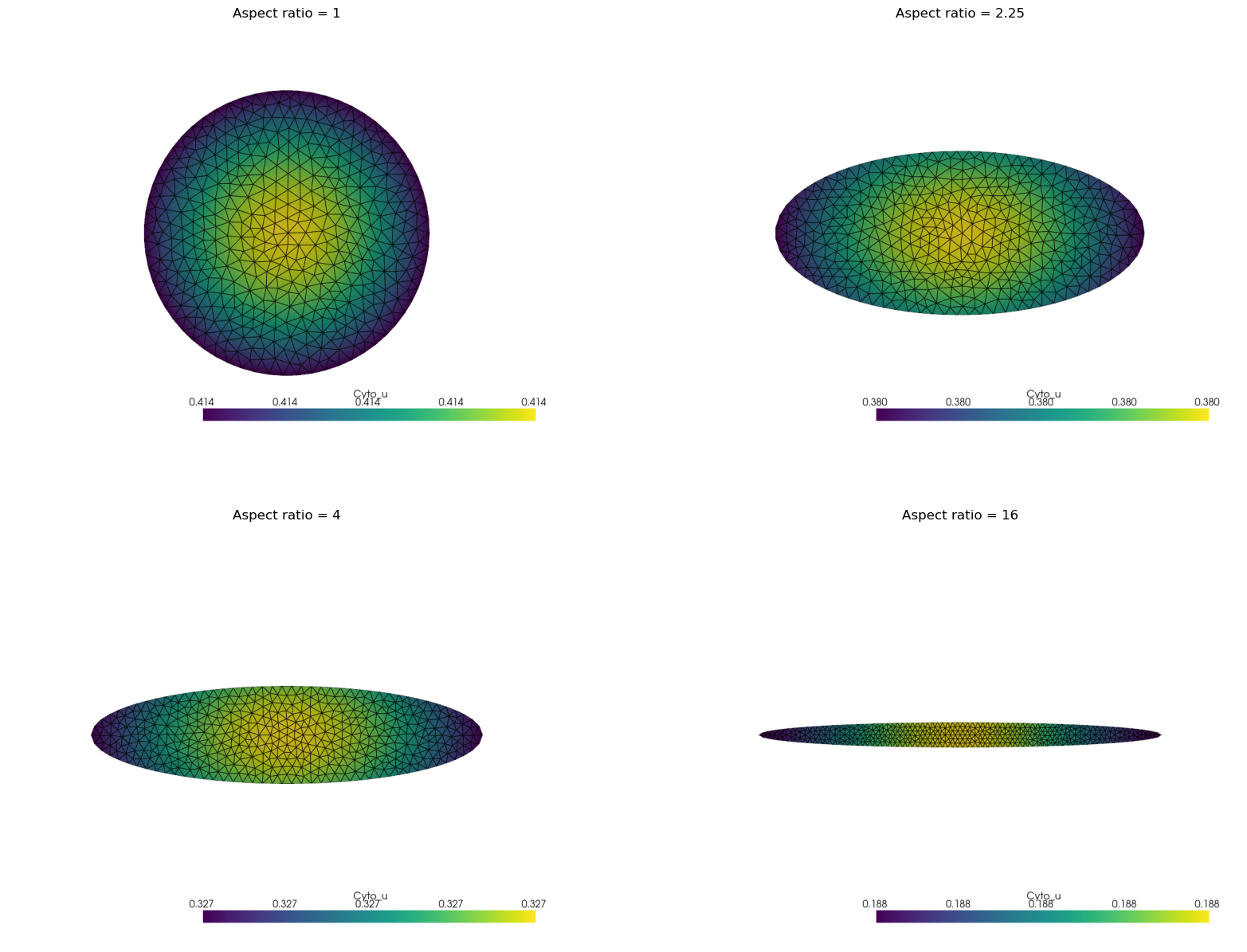
Now iterate over three different cell aspect ratios and compare dynamics between cases.
# initialize plot to compare dynamics
plt.plot(tvec, avg_A, label='Aspect ratio = 1')
plt.xlabel('Time (s)')
plt.ylabel('A concentration $\mathrm{(molecules/μm^2)}$')
# iterate over additional aspect ratios, enforcing the same ellipsoid area in all cases
aspect_ratios = [1, 1.5**2, 4, 16]
ss_calc = [avg_A[-1]]
for i in range(1, len(aspect_ratios)):
# Create mesh
xrad = 1.0*np.sqrt(aspect_ratios[i])
yrad = 1.0/np.sqrt(aspect_ratios[i])
ellipse_mesh, mf1, mf2 = mesh_tools.create_ellipses(xrad, yrad, hEdge=h_ellipse,
outer_tag=surf_tag, outer_marker=edge_tag)
mesh_folder = pathlib.Path(f"ellipse_mesh_AR{aspect_ratios[i]}")
mesh_folder.mkdir(exist_ok=True)
mesh_tools.write_mesh(ellipse_mesh, mf1, mf2, mesh_file)
parent_mesh = mesh.ParentMesh(
mesh_filename=str(mesh_file),
mesh_filetype="hdf5",
name="parent_mesh",
)
config_cur = config.Config()
config_cur.solver.update(
{
"final_t": 5.0,
"initial_dt": 0.05,
"time_precision": 6,
}
)
model_cur = model.Model(pc, sc, cc, rc, config_cur, parent_mesh)
model_cur.initialize()
results = dict()
result_folder = pathlib.Path(f"resultsEllipse_AR{aspect_ratios[i]}")
result_folder.mkdir(exist_ok=True)
for species_name, species in model_cur.sc.items:
results[species_name] = d.XDMFFile(
model_cur.mpi_comm_world, str(result_folder / f"{species_name}.xdmf")
)
results[species_name].parameters["flush_output"] = True
results[species_name].write(model_cur.sc[species_name].u["u"], model_cur.t)
avg_A = [A.initial_condition]
dx = d.Measure("dx", domain=model_cur.cc['Cyto'].dolfin_mesh)
volume = d.assemble_mixed(1.0*dx)
while True:
# Solve the system
model_cur.monolithic_solve()
# Save results for post processing
for species_name, species in model_cur.sc.items:
results[species_name].write(model_cur.sc[species_name].u["u"], model_cur.t)
int_val = d.assemble_mixed(model_cur.sc['A'].u['u']*dx)
avg_A.append(int_val / volume)
# End if we've passed the final time
if model_cur.t >= model_cur.final_t:
break
plt.plot(model_cur.tvec, avg_A, label=f"Aspect ratio = {aspect_ratios[i]}")
ss_calc.append(avg_A[-1])
meshimg_file = meshimg_folder / f"ellipse_mesh_AR{aspect_ratios[i]}.png"
images[aspect_ratios[i]] = visualization.plot(model_cur.sc["A"].u["u"], view_xy=True, filename=meshimg_file)
plt.legend()
Info : Meshing 1D...
Info : Meshing curve 1 (Ellipse)
2025-09-03 20:30:29,498 smart.model - WARNING - Warning! Initial L2-norm of compartment PM is 1.184345213443547 (possibly too large). (model.py:1223)
Info : Done meshing 1D (Wall 0.4776s, CPU 0.473736s)
Info : Meshing 2D...
Info : Meshing surface 1 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0379592s, CPU 0.038296s)
Info : 446 nodes 891 elements
Info : Writing 'tmp_ellipse_[1.5, 0.6666666666666666]_[0.0, 0.0]_0/ellipses.msh'...
Info : Done writing 'tmp_ellipse_[1.5, 0.6666666666666666]_[0.0, 0.0]_0/ellipses.msh'
╒════╤═════════════╤═══════╤══════════════════╤═════════════╤══════════════╤════════════════╕
│ │ name │ id │ dimensionality │ num_cells │ num_facets │ num_vertices │
╞════╪═════════════╪═══════╪══════════════════╪═════════════╪══════════════╪════════════════╡
│ 0 │ parent_mesh │ 12925 │ 2 │ 819 │ 1264 │ 446 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 1 │ Cyto │ 12939 │ 2 │ 819 │ 1264 │ 446 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 2 │ PM │ 12946 │ 1 │ 71 │ 71 │ 71 │
╘════╧═════════════╧═══════╧══════════════════╧═════════════╧══════════════╧════════════════╛
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
Info : Meshing 1D...
Info : Meshing curve 1 (Ellipse)
2025-09-03 20:30:36,053 smart.model - WARNING - Warning! Initial L2-norm of compartment PM is 1.3068209611994575 (possibly too large). (model.py:1223)
Info : Done meshing 1D (Wall 0.591241s, CPU 0.589734s)
Info : Meshing 2D...
Info : Meshing surface 1 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0415244s, CPU 0.041932s)
Info : 457 nodes 913 elements
Info : Writing 'tmp_ellipse_[2.0, 0.5]_[0.0, 0.0]_0/ellipses.msh'...
Info : Done writing 'tmp_ellipse_[2.0, 0.5]_[0.0, 0.0]_0/ellipses.msh'
╒════╤═════════════╤═══════╤══════════════════╤═════════════╤══════════════╤════════════════╕
│ │ name │ id │ dimensionality │ num_cells │ num_facets │ num_vertices │
╞════╪═════════════╪═══════╪══════════════════╪═════════════╪══════════════╪════════════════╡
│ 0 │ parent_mesh │ 19985 │ 2 │ 826 │ 1282 │ 457 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 1 │ Cyto │ 19999 │ 2 │ 826 │ 1282 │ 457 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 2 │ PM │ 20006 │ 1 │ 86 │ 86 │ 86 │
╘════╧═════════════╧═══════╧══════════════════╧═════════════╧══════════════╧════════════════╛
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
Info : Meshing 1D...
Info : Meshing curve 1 (Ellipse)
2025-09-03 20:30:42,983 smart.model - WARNING - Warning! Initial L2-norm of compartment Cyto is 1.264749048952121 (possibly too large). (model.py:1223)
2025-09-03 20:30:42,985 smart.model - WARNING - Warning! Initial L2-norm of compartment PM is 1.7886252580265631 (possibly too large). (model.py:1223)
Info : Done meshing 1D (Wall 0.768621s, CPU 0.768669s)
Info : Meshing 2D...
Info : Meshing surface 1 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0545269s, CPU 0.054803s)
Info : 471 nodes 941 elements
Info : Writing 'tmp_ellipse_[4.0, 0.25]_[0.0, 0.0]_0/ellipses.msh'...
Info : Done writing 'tmp_ellipse_[4.0, 0.25]_[0.0, 0.0]_0/ellipses.msh'
╒════╤═════════════╤═══════╤══════════════════╤═════════════╤══════════════╤════════════════╕
│ │ name │ id │ dimensionality │ num_cells │ num_facets │ num_vertices │
╞════╪═════════════╪═══════╪══════════════════╪═════════════╪══════════════╪════════════════╡
│ 0 │ parent_mesh │ 27045 │ 2 │ 778 │ 1248 │ 471 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 1 │ Cyto │ 27059 │ 2 │ 778 │ 1248 │ 471 │
├────┼─────────────┼───────┼──────────────────┼─────────────┼──────────────┼────────────────┤
│ 2 │ PM │ 27066 │ 1 │ 162 │ 162 │ 162 │
╘════╧═════════════╧═══════╧══════════════════╧═════════════╧══════════════╧════════════════╛
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
<matplotlib.legend.Legend at 0x7f9b68e92140>
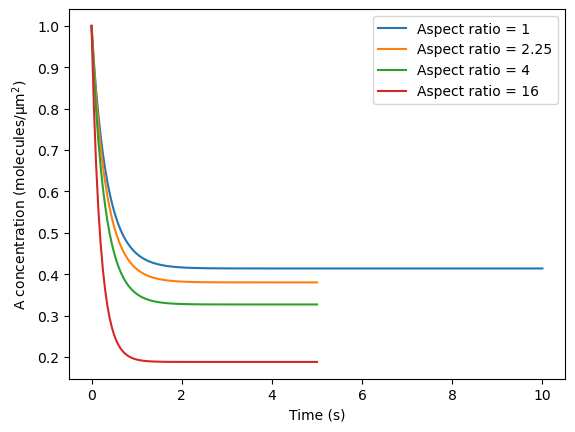
Compare steady-state values with those from past runs using SMART (regression test):
ss_stored = [0.41384929369203755, 0.38021621231324093, 0.32689497127690803, 0.18803994874948224]
percent_error = max(np.abs(np.array(ss_stored) - np.array(ss_calc))/np.array(ss_stored))*100
assert percent_error < .01,\
f"Failed regression test: Example 2 results deviate {percent_error:.3f}% from the previous numerical solution"
Now, display the final states from all simulations for direct comparison.
fig, ax = plt.subplots(2, 2, figsize=(20, 15))
for axi, (aspect_ratio, image) in zip(ax.flatten(), images.items()):
axi.imshow(image)
axi.axis('off')
axi.set_title(f"Aspect ratio = {aspect_ratio}")